Deep Learning Architectures
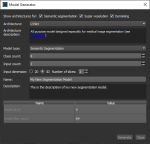
The Model Generator dialog for the Deep Learning Tool and the Segmentation Wizard provides options for generating new Deep Learning models with a number of different architectures.
Click the New button on the Model Overview panel in the Deep Learning Tool or on the Models tab on the Segmentation Wizard panel to open the Model Generator dialog, shown below.
Model Generator dialog for the Deep Learning Tool
The following table lists the deep learning architectures available for semantic segmentation, super-resolution, and denoising. Note that the Segmentation Wizard only provides options for selecting architectures implemented for semantic segmentation.
|
|
User for |
Description |
|---|---|---|
|
Attention U-Net |
Semantic segmentation |
This attention gate (AG) model, which was originally designed for medical imaging segmentation, automatically learns to focus on target structures of varying shapes and sizes while suppressing irrelevant regions in input images. By highlighting salient features only, the necessity of using explicit external tissue/organ localization module of cascaded convolutional neural networks (CNNs) is eliminated. Integrated into the standard U-Net architecture, AGs can increase model sensitivity and prediction accuracy. Reference… Oktay et al. Attention U-Net: Learning Where to Look for the Pancreas, arXiv, May 20, 2018 (also available online at https://arxiv.org/pdf/1804.03999.pdf). |
|
Auto-Encoder |
Semantic segmentation, denoising |
Generic autoencoder. Reference… Additional information about Autoencoder is available online at: https://en.wikipedia.org/wiki/Autoencoder. |
|
DeepLabV3+ |
Semantic segmentation |
Reference… Chen et al. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation, arXiv, August 22, 2018 (also available online at https://arxiv.org/pdf/1802.02611.pdf). |
|
EDSR |
Super-resolution |
Reference… Lim et el. Enhanced Deep Residual Networks for Singe Image Super-Resolution, arXiv, July 10, 2017 (also available online at https://arxiv.org/pdf/1707.02921.pdf). |
|
FC-DenseNet |
Semantic segmentation |
Reference… Jégou et al. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation, arXiv, October 31, 2017 (also available online at https://arxiv.org/pdf/1611.09326.pdf). |
|
LinkNet |
Semantic segmentation |
This architecture focuses on speed and efficiency for semantic segmentation tasks. Compared to other algorithms, LinkNet can learn with a more limited number of parameters and operations and still deliver accurate results. Reference… Abhishek Chaurasia and Eugenio Culurciello, LinkNet: Exploiting Encoder Representations for Efficient Semantic Segmentation, arXiv, June 14, 2017 (also available online at https://arxiv.org/pdf/1611.09326.pdf). |
|
Noise2Noise |
Denoising |
Reference… Lehtinen et al. Noise2Noise: Learning Image Restoration without Clean Data, arXiv, October 29, 2018 (also available online at https://arxiv.org/pdf/1803.04189.pdf). |
|
Noise2Noise_SRResNet |
Denoising |
Reference… Lehtinen et al. Noise2Noise: Learning Image Restoration without Clean Data, arXiv, October 29, 2018 (also available online at https://arxiv.org/pdf/1803.04189.pdf). |
|
PSPNet |
Semantic segmentation |
Reference… Zhao et al. Pyramid Scene Parsing Network, arXiv, April 27, 2017 (also available online at: https://arxiv.org/pdf/1612.01105.pdf). |
|
Sensor3D |
Semantic segmentation |
Semantic segmentation model using convolution LSTM. Reference… Novikov et al. Deep Sequential Segmentation of Organs in Volumetric Medical Scans, arXiv, March 11, 2019 (also available online at https://arxiv.org/pdf/1807.02437.pdf). |
|
U-Net |
Semantic segmentation, super-resolution, denoising |
All purpose model designed especially for medical image segmentation. Reference… Ronneberger et al. U-Net: Convolutional Networks for Biomedical Image Segmentation, arXiv, May 18, 2015 (also available online at https://arxiv.org/pdf/1505.04597.pdf). |
|
U-Net 3D |
Semantic segmentation, super-resolution, denoising |
3D implementation of U-Net. Reference… Ronneberger et al. U-Net: Convolutional Networks for Biomedical Image Segmentation, arXiv, May 18, 2015 (also available online at https://arxiv.org/pdf/1505.04597.pdf). Note Currently, only U-Net 3D is a fully 3D model that uses 3D convolutions. The number of input slices for this model is determined by the input size, which must be cubic. For example, 32x32x32. U-Net uses 2D convolutions, but can take 3D input patches for which you can choose the number of slices. You should also note that in some cases, 3D models can be more reliable for segmentation tasks. |
|
U-Net++ |
Semantic segmentation, super-resolution, denoising |
U-Net++ is a powerful architecture for medical image and semantic segmentation. This architecture is a deeply-supervised encoder-decoder network in which the encoder and decoder sub-networks are connected through a series of nested, dense skip pathways. The skip pathways help reduce the semantic gap between the feature maps of the encoder and decoder sub-networks. Reference… Zhou et al. UNet++: A Nested U-Net Architecture for Medical Image Segmentation, arXiv, July 18, 2018. |
|
WDSR |
Super-resolution |
Reference… Yu et al. Wide Activation for Efficient and Accurate Image Super-Resolution, arXiv, December 21, 2018 (also available online at https://arxiv.org/pdf/1808.08718.pdf). |
A limited number of the basic parameters for Deep Learning architectures are available for editing in the Model Generator. These are listed below.
Refer to the documents referenced in the table above for more information about the editable parameters available for the implemented architectures.
|
|
Description |
|---|---|
|
Attention U-Net |
Depth level… Depth of the network, as determined by the number of pooling layers. Initial filter count… Filter count at the first convolution layer. |
|
Autoencoder |
Initial filter count… Filter count at the first convolutional layer. Kernel size… Convolutional filters kernel size. Pooling size… Pooling window size. |
|
BiSeNet |
Patch size… Size of the input patches. |
|
DeepLabV3+ |
Backbone… Backbone to use — 'Xception' or 'MobileNetV2'. Patch size… Fixed size of the input patches. Output stride… Ratio of the image size to the encoder output size. |
|
EDSR |
Scale… Ratio of the input size to the output size. Patch size… Fixed size of the input patches. Filter count… Filter count at each convolution layer. ResNet block count… The number of times to repeat ResNet blocks. Use Tanh activation… Determines if Tanh activation will be applied — True or False. |
|
FC-DenseNet |
Model type… Model variation to be generated — FC-DenseNet56, FC-DenseNet67, or FC-DenseNet103. |
|
LinkNet |
Patch size… Fixed size of the input patches. Initial filter count… Filter count at the first convolution layer. |
|
Noise2Noise |
Initial filter count… Filter count at the first convolution layer. |
|
Noise2Noise_SRResNet |
Filter count… Filter count at each convolution layer. ResNet block count… The number of times to repeat ResNet blocks. |
|
PSPNet |
Backbone… Backbone to use — ResNet50 or ResNet101. Patch size… Fixed size of the input patches. Filter count… Filter count at each convolution layer. |
|
Sensor3D |
Depth level… Depth of the network, as determined by the number of pooling layers, and patch size. Initial filter count… Filter count at the first convolution layer. |
|
U-Net |
Depth level… Depth of the network, as determined by the number of pooling layers. Initial filter count… Filter count at the first convolution layer. |
|
U-Net 3D |
Topology… The topology of the model. Initial filter count… Filter count at the first convolution layer. Use batch normalization… Determines if batch normalization will be applied — True or False. |
|
U-Net++ |
Depth level… Depth of the network, as determined by the number of pooling layers. Initial filter count… Filter count at the first convolution layer. |
|
WDSR |
Model type… Model variation to be generated — WDSR-A or WDSR-B. Scale… Ratio of the input size to the output size. Patch size…Fixed size of the input patches. Filter count… Filter count at each convolution layer. ResNet block count… The number of times to repeat ResNet blocks. ResNet block expansion… The ratio to multiple the number of filters in the ResNet block expansion layer. |